-Search query
-Search result
Showing 1 - 50 of 85 items for (author: kaelber & jt)

EMDB-41637: 
Asymmetric cryoEM reconstruction of Mayaro virus
Method: single particle / : Chmielewski D, Kaelber J, Jin J, Weaver S, Auguste AJ, Chiu W

EMDB-28979: 
Cryo-EM structure of Chikungunya virus asymmetric unit
Method: single particle / : Su GC, Chmielewsk D, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-41096: 
Cryo-electron tomography of Chikungunya virus pentamer structure
Method: subtomogram averaging / : Chmielewsk D, Su GC, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-41631: 
Cryo-EM structure of Chikungunya virus with asymmetric reconstruction
Method: single particle / : Su GC, Chmielewsk D, Kaelber J, Pintilie G, Chen M, Jin J, Auguste A, Chiu W

EMDB-41125: 
Zophobas morio black wasting virus strain NJ2-molitor virion structure
Method: single particle / : Penzes JJ, Kaelber JT
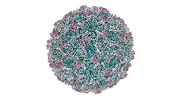
EMDB-41106: 
Zophobas morio black wasting virus strain UT-morio virion structure
Method: single particle / : Penzes JJ, Kaelber JT
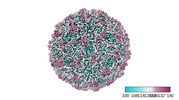
EMDB-41301: 
Zophobas morio black wasting virus strain OR-molitor virion structure
Method: single particle / : Penzes JJ, Kaelber JT
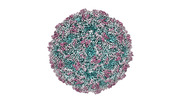
EMDB-41108: 
Zophobas morio black wasting virus strain UT-morio empty capsid structure
Method: single particle / : Penzes JJ, Kaelber JT
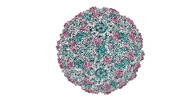
EMDB-41131: 
Zophobas morio black wasting virus strain OR-molitor empty capsid structure
Method: single particle / : Penzes JJ, Kaelber JT

EMDB-33403: 
3.1 Angstrom cryoEM icosahedral reconstruction of mud crab reovirus
Method: single particle / : Zhang Q, Gao Y

EMDB-33404: 
3.4 Angstrom cryoEM D5 reconstruction of mud crab reovirus
Method: single particle / : Zhang QF, Gao YZ

EMDB-33405: 
icosahedral reconstruction of mud crab reovirus in transcriptionally active state
Method: single particle / : Zhang Q, Gao Y

EMDB-33406: 
D5 reconstruction of mud crab reovirus in transcriptionally active state
Method: single particle / : Zhang Q, Gao Y

EMDB-28780: 
Eilat virus/Eastern equine encephalitis virus chimeric vaccine candidate
Method: single particle / : Kaelber JT, Chmielewski D, Chiu W, Auguste AJ

EMDB-26438: 
Transcription antitermination complex: "pre-engaged" Qlambda-loading complex
Method: single particle / : Yin Z, Ebright RH

EMDB-26439: 
Transcription antitermination complex: NusA-containing "engaged" Qlambda-loading complex
Method: single particle / : Yin Z, Ebright RH

EMDB-27864: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, Mg-ADP-BeF3, and NusG; TEC part
Method: single particle / : Molodtsov V, Wang C

EMDB-27865: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part
Method: single particle / : Molodtsov V, Wang C

EMDB-27897: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, Mg-ADP-BeF3, and NusG
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-27913: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 21 nt long RNA spacer, Mg-ADP-BeF3, and NusG; TEC part
Method: single particle / : Molodtsov V, Wang C

EMDB-27914: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 21 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part
Method: single particle / : Molodtsov V, Wang C

EMDB-27915: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 21 nt long RNA spacer, Mg-ADP-BeF3, and NusG
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-27916: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 24 nt long RNA spacer, Mg-ADP-BeF3, and NusG; TEC part
Method: single particle / : Molodtsov V, Wang C

EMDB-27917: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 24 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part
Method: single particle / : Molodtsov V, Wang C

EMDB-27918: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 24 nt long RNA spacer, Mg-ADP-BeF3, and NusG
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-27928: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, lambda-tR1 rut RNA, Mg-ADP-BeF3, and NusG; Rho part
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-27929: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, lambda-tR1 rut RNA, Mg-ADP-BeF3, and NusG
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-27930: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, lambda-tR1 rut RNA, Mg-ADP-BeF3, and NusG; TEC part
Method: single particle / : Molodtsov V, Wang C

EMDB-27931: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, dC75 rut mimic RNA, Mg-ADP-BeF3, and NusG; TEC part
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-27932: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, dC75 rut mimic RNA, Mg-ADP-BeF3, and NusG; Rho hexamer part
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-27933: 
Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, dC75 rut mimic RNA
Method: single particle / : Molodtsov V, Wang C, Ebright RH

EMDB-23378: 
CryoEM structure of Mayaro virus
Method: single particle / : Chmielewski D, Kaelber J

EMDB-24424: 
Cryo-EM structure of Thermus thermophilus reiterative transcription complex with 11nt oligo-G RNA
Method: single particle / : Liu Y, Ebright RH

EMDB-21959: 
Colicin E1 fragment in nanodisc-embedded TolC
Method: single particle / : Kaelber JT, Budiardjo SJ, Firlar E, Case DA, Ikujuni AP, Slusky JSG

EMDB-21960: 
Colicin E1 fragment in nanodisc-embedded TolC
Method: single particle / : Kaelber JT, Budiardjo SJ, Firlar E, Ikujuni AP, Slusky JSG

EMDB-22341: 
CryoEM structure of Streptococcus thermophilus SHP pheromone receptor Rgg3 in complex with SHP3
Method: single particle / : Petrou VI, Capodagli GC

EMDB-22412: 
Adeno-associated virus strain AAV7 capsid icosahedral structure
Method: single particle / : Firlar E, Yost SA

EMDB-21670: 
Large ribosomal subunit of Halococcus morrhuae, an archaeon with an unusual 5S rRNA insertion
Method: single particle / : Tirumalai MR, Kaelber JT, Fox GE

EMDB-21386: 
Cryo-EM structure of Escherichia coli transcription-translation complex A (TTC-A) containing mRNA with a 12 nt long spacer
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright R

EMDB-21468: 
Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 15 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21469: 
Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 18 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21470: 
Escherichia coli transcription-translation complex A1 (TTC-A1) containing a 21 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21471: 
Escherichia coli transcription-translation complex A2 (TTC-A2) containing a 15 nt long mRNA spacer, NusG, and fMet-tRNAs at P-site and E-site
Method: single particle / : Molodtsov V, Wang C, Ebright RH, Su M

EMDB-21472: 
Escherichia coli transcription-translation complex C2 (TTC-C2) containing a 27 nt long mRNA spacer
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21474: 
Escherichia coli transcription-translation complex C3 (TTC-C3) containing mRNA with a 27 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21475: 
Escherichia coli transcription-translation complex C4 (TTC-C4) containing mRNA with a 21 nt long spacer, transcription factor NusG, and fMet-tRNAs at P-site and E-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21476: 
Escherichia coli transcription-translation complex C5 (TTC-C5) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21477: 
Escherichia coli transcription-translation complex C6 (TTC-C6) containing mRNA with a 21 nt long spacer, NusA, and fMet-tRNAs at E-site and P-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21482: 
Escherichia coli transcription-translation complex D1 (TTC-D1) containing mRNA with a 27 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH

EMDB-21483: 
Escherichia coli transcription-translation complex D2 (TTC-D2) containing mRNA with a 27 nt long spacer
Method: single particle / : Molodtsov V, Wang C, Su M, Ebright RH
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
